MetDisease Plugin for Cytoscape
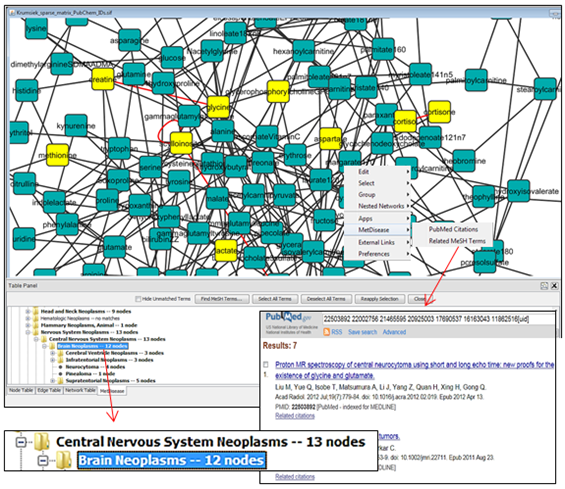
MetDisease is an app for Cytoscape, the bioinformatics network visualization tool. The app is used to annotate a metabolic network with MeSH disease terms, explore related diseases within a network, and link to PubMed references corresponding to any network node and selection of MeSH terms. MeSH terms are controlled vocabulary terms used by the National Library of Medicine to describe the content of the articles indexed in PubMed.
Download the latest version of MetDisease | Download MetDisease user manual
Download sample input files: Fig_1_network.xgmml | Suppl_fig_1_network.xgmml
PI
Developers
Bill Duren
Tim Hull
Terry Weymouth
Acknowledgments
We thank Marci Brandenburg For creating MetDIsease user manual
Funding
This work is supported by NIH Grants
U54DA021519 and U24DK097153
Please report any problems to metscape-help@umich.edu
